Julia Transformation
Note: This feature is in preview and available on request via our support.
Julia transformations complement the R and SQL transformations where computations or other operations are too difficult. Common data operations like joining, sorting, or grouping are still easier and faster to do in the SQL Transformations.
Environment
The Julia script is running in an isolated Docker environment. The current Julia version is 1.5.0. We release updates regularly, few weeks after the official release, announcing them on our status page.
When we update a Julia version, we offer — for a limited time — the option to switch to the previous version. You can
switch the version in the transformation detail by clicking on the Backend Version label:
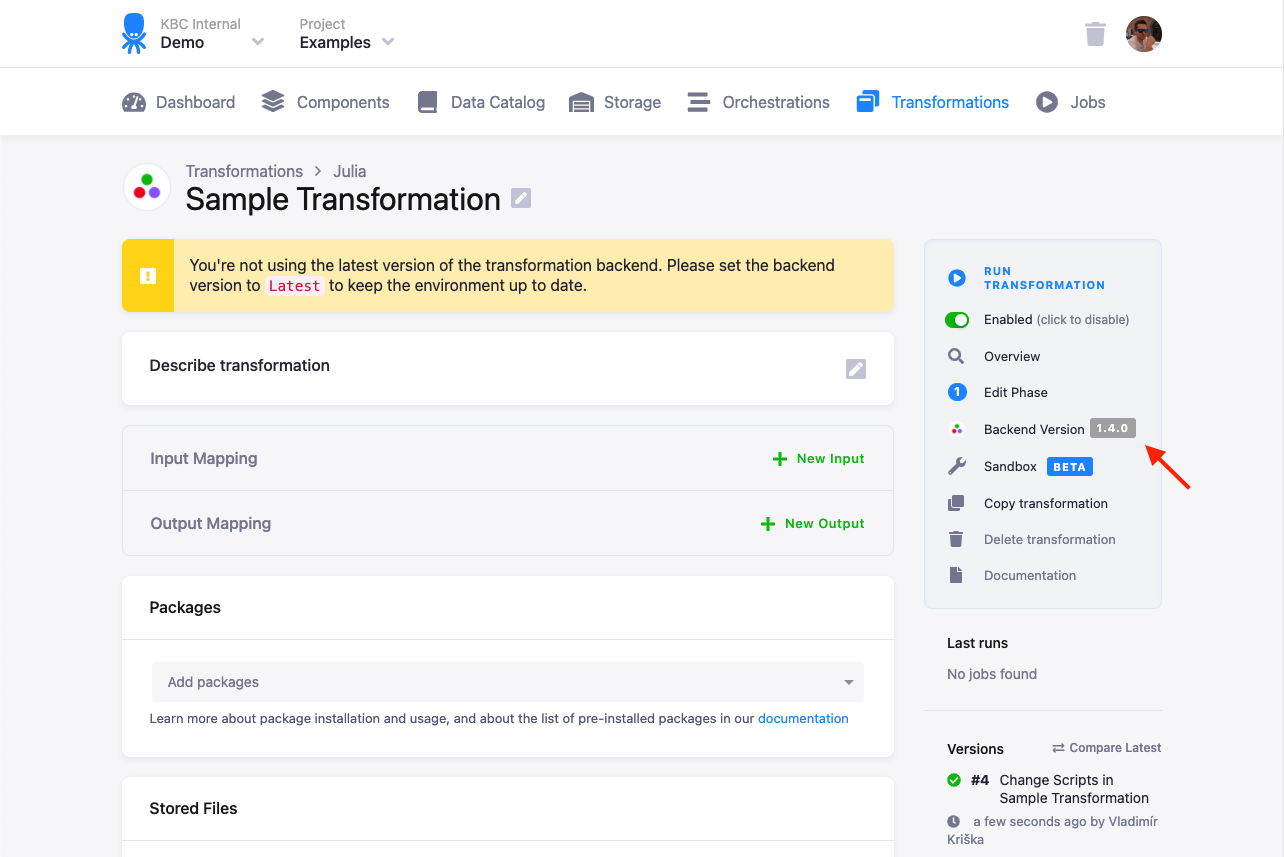
This feature is intended to help you ‘postpone’ the update to a more convenient time for you in case there are any problems with the new version. You should update the transformation code to the new version soon, as the old version is considered unsupported.
If the Julia label is not displayed, there is no previous version offered. It is still possible to change the version
via the API though.
Memory and Processing Constraints
The Docker container running the Julia transformation has 8GB of allocated memory, and the maximum running time is 6 hours. The container is also limited to the equivalent of two Intel Broadwell 2.3 GHz processors.
File Locations
The Julia script itself will be compiled to /data/script.jl. To access your input and output tables, use
relative (in/tables/file.csv, out/tables/file.csv) or absolute (/data/in/tables/file.csv, /data/out/tables/file.csv) paths.
To access downloaded files, use the in/files/ or /data/in/files/ path. If you want to dig really deep,
have a look at the full Common Interface specification.
Temporary files can be written to a /tmp/ folder. Do not use the /data/ folder for files you do not wish to exchange with Keboola.
Packages
You can list extra packages in the UI. These packages are installed using the General package registry.
Normally, any listed package can be installed. However, some packages have external dependencies that might not be available.
Feel free to contact us if you run into problems. When a package is installed, you still need to have a using or import statement in your script.
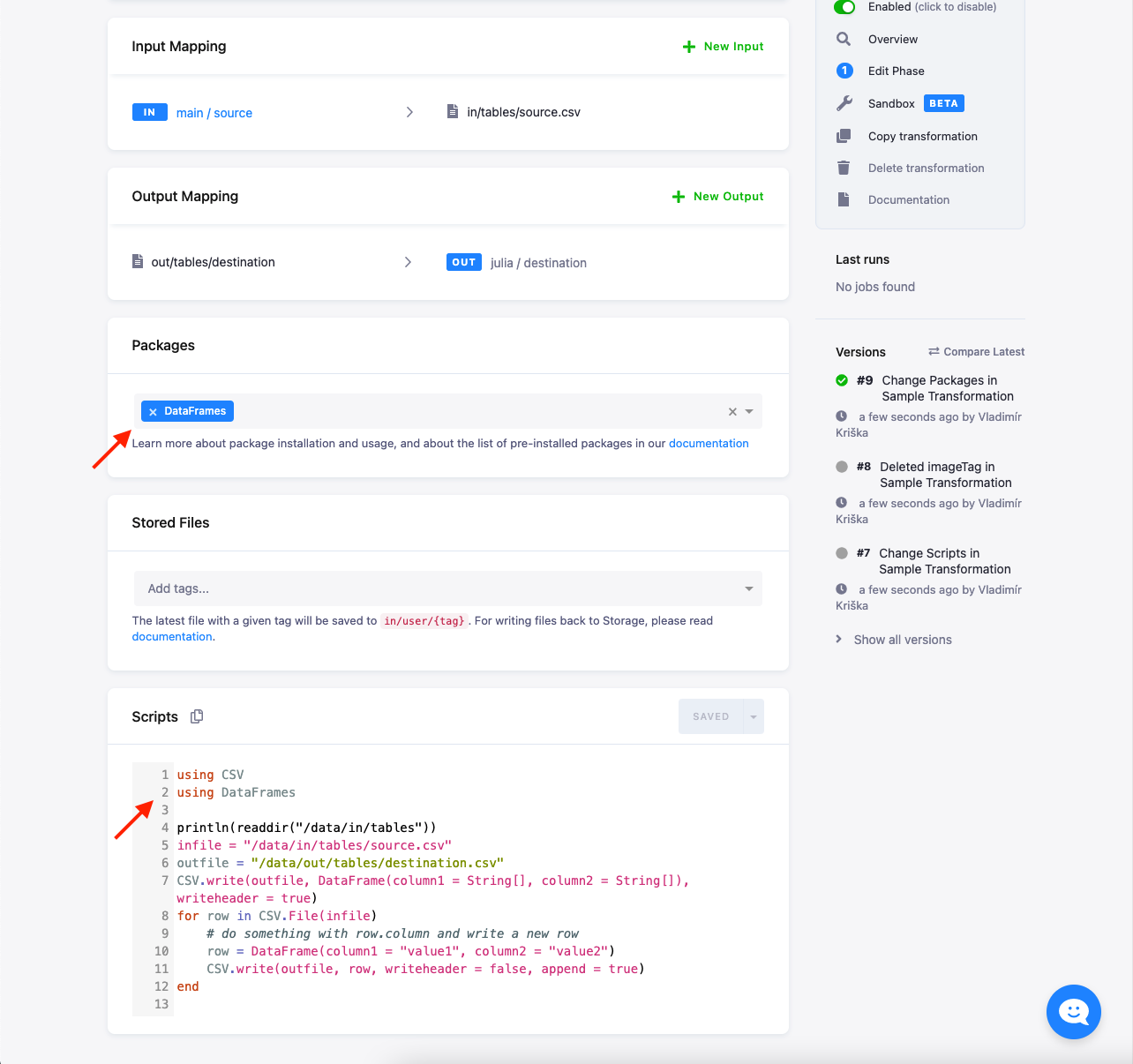
The latest versions of packages are always installed at the time of the release (you can check that in the repository). In case your code relies on a specific package version, you can override the installed version by calling, e.g.:
using Pkg
Pkg.add(PackageSpec(name="Example", version="0.3"))Some packages are already installed in the environment (see the full list); they do not need to be listed in the transformation.
CSV Format
Tables from Storage are imported to the Julia script from CSV files. CSV files can be read by functions
from the CSV package.
You can read CSV files either to vectors (numbered columns), or to dictionaries (named columns).
Your input tables are stored as CSV files in in/tables/ and your output tables are stored in out/tables/.
If you can process the file line-by-line, then the most effective way is to read each line, process it and write it immediately. The following example shows how to read and manipulate a CSV file:
using CSV
using DataFrames
infile = "in/tables/source.csv"
outfile = "out/tables/result.csv"
CSV.write(outfile, DataFrame(column1 = String[], column2 = String[]), writeheader = true)
for row in CSV.File(infile)
# do something with row.column and write a new row
row = DataFrame(column1 = row.first * "ping", column2 = row.second * 42)
CSV.write(outfile, row, writeheader = false, append = true)
endLocal Development Tutorial
To develop and debug Julia transformations, we recommend that you use the Sandbox feature. It gives you an environment very similar to the transformations out of the box.
If you want to develop the transformation code on your local machine, you can do so by replicating the execution environment. You just need to have Julia installed, preferably the same version as us.
All you need to do to simulate the input and output mapping is create the right directories with the right files. The following image shows the directory structure:
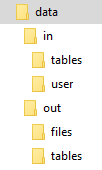
The script itself is expected to be in the data directory; its name is arbitrary. It is possible to use relative directories
so that you can move the script to a transformation with no changes. To develop a Julia transformation that takes a sample CSV file locally, follow these steps:
- Put the Julia code into a file, for example, script.jl, in the working directory.
- Put all the input mapping tables inside the
in/tablessubdirectory of the working directory. - If using binary files, place them inside the
in/usersubdirectory of the working directory, and make sure that their name has no extension. - Store the result CSV files inside the
out/tablessubdirectory.
Use the above sample script. A finished example of the script is attached below in data.zip.
Download it and test the script in your local Julia installation. The output file result.csv will be created.
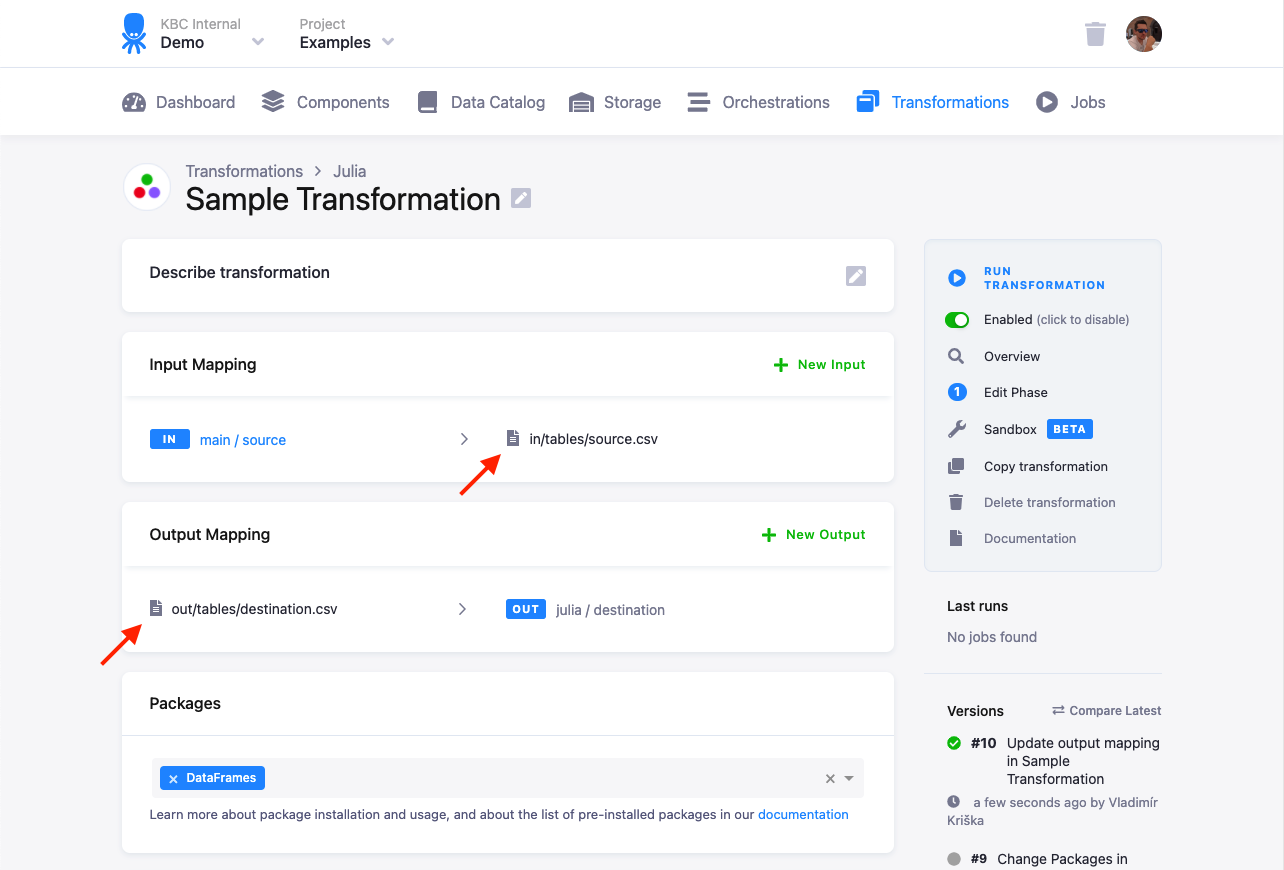
This script can be used in your transformations without any modifications. All you need to do is
- create a table in Storage by uploading the sample CSV file),
- create an input mapping from that table, setting its destination to
source.csv(as expected by the Julia script), - create an output mapping from
result.csv(produced by the Julia script) to a new table in your Storage, - copy & paste the above script into the transformation code, and finally,
- save and run the transformation.
Going Further
The above steps are usually sufficient for daily development and the debugging of moderately complex Julia transformations, although they do not reproduce the transformation execution environment exactly. To create a development environment with the exact same configuration as the transformation environment, use our Docker image.
